Redun¶
Here, we’ll see how to track redun workflow runs with LaminDB.
Note
This use case is based on github.com/ricomnl/bioinformatics-pipeline-tutorial.
!lamin init --storage ./test-redun-lamin --schema bionty
Show code cell output
Operations to perform:
Apply all migrations: bionty, contenttypes, lamindb
Running pre-migrate handlers for application contenttypes
Running pre-migrate handlers for application lamindb
Running pre-migrate handlers for application bionty
Running migrations:
Applying lamindb.0069_squashed...
OK (1.982s)
Applying lamindb.0070_lamindbv1_migrate_data...
OK (1.193s)
Applying lamindb.0071_lamindbv1_migrate_schema...
OK (4.951s)
Applying lamindb.0072_remove_user__branch_code_remove_user_aux_and_more...
OK (0.798s)
Applying lamindb.0073_merge_ourprojects...
OK (1.393s)
Applying lamindb.0074_lamindbv1_part4...
OK (4.158s)
Applying bionty.0041_squashed...
OK (11.058s)
Applying bionty.0042_lamindbv1...
OK (6.628s)
Applying bionty.0043_lamindbv2_part2...
OK (2.172s)
Applying bionty.0044_alter_cellline_space_alter_cellmarker_space_and_more...
OK (2.291s)
Applying bionty.0045_rename_aux_cellline__aux_rename_aux_cellmarker__aux_and_more...
OK (2.268s)
Applying bionty.0046_alter_cellline__aux_alter_cellmarker__aux_and_more...
OK (2.271s)
Applying lamindb.0075_lamindbv1_part5...
OK (5.580s)
Applying bionty.0047_lamindbv1_part5...
OK (3.227s)
Applying contenttypes.0001_initial... OK (0.011s)
Applying contenttypes.0002_remove_content_type_name...
OK (0.157s)
Running post-migrate handlers for application contenttypes
Adding content type 'contenttypes | contenttype'
Running post-migrate handlers for application lamindb
Adding content type 'lamindb | feature'
Adding content type 'lamindb | param'
Adding content type 'lamindb | user'
Adding content type 'lamindb | artifact'
Adding content type 'lamindb | collection'
Adding content type 'lamindb | collectionartifact'
Adding content type 'lamindb | featurevalue'
Adding content type 'lamindb | artifactfeaturevalue'
Adding content type 'lamindb | paramvalue'
Adding content type 'lamindb | artifactparamvalue'
Adding content type 'lamindb | run'
Adding content type 'lamindb | runparamvalue'
Adding content type 'lamindb | storage'
Adding content type 'lamindb | transform'
Adding content type 'lamindb | ulabel'
Adding content type 'lamindb | collectionulabel'
Adding content type 'lamindb | artifactulabel'
Adding content type 'lamindb | space'
Adding content type 'lamindb | transformulabel'
Adding content type 'lamindb | artifactproject'
Adding content type 'lamindb | artifactreference'
Adding content type 'lamindb | collectionproject'
Adding content type 'lamindb | collectionreference'
Adding content type 'lamindb | person'
Adding content type 'lamindb | project'
Adding content type 'lamindb | reference'
Adding content type 'lamindb | transformproject'
Adding content type 'lamindb | transformreference'
Adding content type 'lamindb | schema'
Adding content type 'lamindb | artifactschema'
Adding content type 'lamindb | schemafeature'
Adding content type 'lamindb | schemaparam'
Running post-migrate handlers for application bionty
Adding content type 'bionty | artifactcellline'
Adding content type 'bionty | artifactcellmarker'
Adding content type 'bionty | artifactcelltype'
Adding content type 'bionty | artifactdevelopmentalstage'
Adding content type 'bionty | artifactdisease'
Adding content type 'bionty | artifactethnicity'
Adding content type 'bionty | artifactexperimentalfactor'
Adding content type 'bionty | artifactgene'
Adding content type 'bionty | artifactorganism'
Adding content type 'bionty | artifactpathway'
Adding content type 'bionty | artifactphenotype'
Adding content type 'bionty | artifactprotein'
Adding content type 'bionty | artifacttissue'
Adding content type 'bionty | cellline'
Adding content type 'bionty | cellmarker'
Adding content type 'bionty | celltype'
Adding content type 'bionty | developmentalstage'
Adding content type 'bionty | disease'
Adding content type 'bionty | ethnicity'
Adding content type 'bionty | experimentalfactor'
Adding content type 'bionty | gene'
Adding content type 'bionty | organism'
Adding content type 'bionty | pathway'
Adding content type 'bionty | phenotype'
Adding content type 'bionty | protein'
Adding content type 'bionty | source'
Adding content type 'bionty | tissue'
Adding content type 'bionty | schemacellmarker'
Adding content type 'bionty | schemagene'
Adding content type 'bionty | schemapathway'
Adding content type 'bionty | schemaprotein'
→ initialized lamindb: testuser1/test-redun-lamin
Amend the workflow¶
import lamindb as ln
import json
→ connected lamindb: testuser1/test-redun-lamin
Let’s amend a redun workflow.py to register input & output artifacts in LaminDB:
To track the workflow run in LaminDB, add (see on GitHub):
ln.track(params=params)
To register the output file via LaminDB, add (see on GitHub):
ln.Artifact(output_path, description="results").save()
Run redun¶
Let’s see what the input files are:
!ls ./fasta
KLF4.fasta MYC.fasta PO5F1.fasta SOX2.fasta
And call the workflow:
!redun run workflow.py main --input-dir ./fasta --tag run=test-run 1> redun_stdout.txt 2>redun_stderr.txt
Inspect the output:
!cat redun_stdout.txt
→ connected lamindb: testuser1/test-redun-lamin
→ running outside of synched git repo, cloning https://github.com/laminlabs/redun-lamin into /home/runner/.cache/lamindb/redun-lamin
! code blob hash e37260d21609885c5a16a79fe01de24b0f074df7 was found in non-default branch(es): origin/slacknotif
→ created Transform('taasWKawCiNA0000'), started new Run('EDruK7Uz...') at 2025-01-14 21:01:21 UTC
! git repo /home/runner/.cache/lamindb/redun-lamin already exists locally
! code blob hash e37260d21609885c5a16a79fe01de24b0f074df7 was found in non-default branch(es): origin/slacknotif
→ created Transform('taasWKawCiNA0000'), started new Run('AjDvah2V...') at 2025-01-14 21:01:22 UTC
→ params: input_dir=./fasta, amino_acid=C, enzyme_regex=[KR], missed_cleavages=0, min_length=4, max_length=75, executor=Executor.default
! folder is outside existing storage location, will copy files from ./fasta to /home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/fasta
?25l
downloading... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0% -:--:--
downloading... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━ 74% 0:00:01
downloading... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00
downloading... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00
?25h
→ finished Run('AjDvah2V') after 0d 0h 0m 6s at 2025-01-14 21:01:28 UTC
File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=a5ecf173)
And the error log:
!tail -1 redun_stderr.txt
[redun] Execution duration: 12.66 seconds
View data lineage:
artifact = ln.Artifact.filter(description="results", suffix=".tgz").one()
artifact.view_lineage()
Track the redun execution id¶
If we want to be able to query LaminDB for redun execution ID, this here is a way to get it:
# export the run information from redun
!redun log --exec --exec-tag run=test-run --format json --no-pager > redun_exec.json
# load the redun execution id from the JSON and store it in the LaminDB run record
with open("redun_exec.json") as file:
redun_exec = json.loads(file.readline())
artifact.run.reference = redun_exec["id"]
artifact.run.reference_type = "redun_id"
artifact.run.save()
Run(uid='AjDvah2V8Xw60bTrOkCv', started_at=2025-01-14 21:01:22 UTC, finished_at=2025-01-14 21:01:28 UTC, reference='70307f22-9649-4042-b6b7-e7de177ab1bf', reference_type='redun_id', space_id=1, transform_id=1, report_id=6, environment_id=5, created_by_id=1, created_at=2025-01-14 21:01:22 UTC)
Track the redun run report¶
Attach a run report:
report = ln.Artifact(
"redun_stderr.txt",
description=f"Redun run report of {redun_exec['id']}",
run=False,
visibility=0,
).save()
artifact.run.report = report
artifact.run.save()
Run(uid='AjDvah2V8Xw60bTrOkCv', started_at=2025-01-14 21:01:22 UTC, finished_at=2025-01-14 21:01:28 UTC, reference='70307f22-9649-4042-b6b7-e7de177ab1bf', reference_type='redun_id', space_id=1, transform_id=1, report_id=8, environment_id=5, created_by_id=1, created_at=2025-01-14 21:01:22 UTC)
View transforms and runs in LaminHub¶
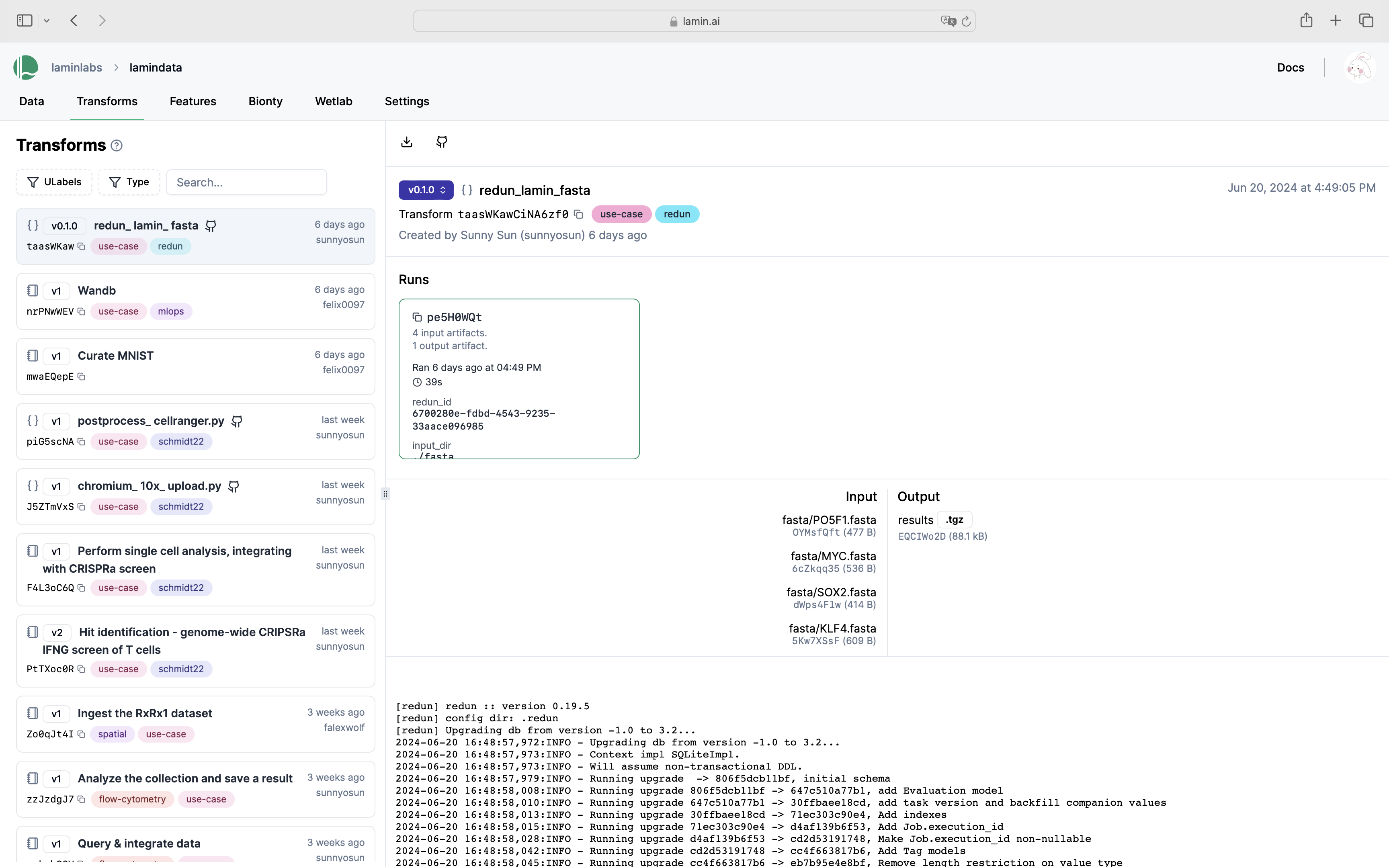
View the database content¶
ln.view()
****************
* module: core *
****************
Artifact
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | _hash_type | _key_is_virtual | _overwrite_versions | space_id | storage_id | schema_id | version | is_latest | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||||
| 7 | A1EEzDtCtlQHvFNn0000 | data/results.tgz | results | .tgz | None | None | 83556 | BCOM2cx_DSzQn4Wu4Ip6dA | None | None | md5 | False | False | 1 | 1 | None | None | True | 2.0 | 2025-01-14 21:01:33.101173+00:00 | 1 | None | 1 |
| 4 | Hr8tju9S02Qz88vL0000 | fasta/PO5F1.fasta | None | .fasta | None | None | 477 | -7iJgveFO9ia0wE1bqVu6g | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-01-14 21:01:23.959035+00:00 | 1 | None | 1 |
| 3 | mfDXA89XArRzSTIw0000 | fasta/SOX2.fasta | None | .fasta | None | None | 414 | C5q_yaFXGk4SAEpfdqBwnQ | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-01-14 21:01:23.958471+00:00 | 1 | None | 1 |
| 2 | esRNQNhzhAeJt7o50000 | fasta/KLF4.fasta | None | .fasta | None | None | 609 | LyuoYkWs4SgYcH7P7JLJtA | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-01-14 21:01:23.957725+00:00 | 1 | None | 1 |
| 1 | blGLxbZyHJpDjSWn0000 | fasta/MYC.fasta | None | .fasta | None | None | 536 | WGbEtzPw-3bQEGcngO_pHQ | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-01-14 21:01:23.956527+00:00 | 1 | None | 1 |
Param
| name | dtype | is_type | _expect_many | space_id | type_id | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||
| 7 | executor | str | None | False | 1 | None | None | 2025-01-14 21:01:20.570411+00:00 | 1 | None | 1 |
| 6 | max_length | int | None | False | 1 | None | None | 2025-01-14 21:01:20.570343+00:00 | 1 | None | 1 |
| 5 | min_length | int | None | False | 1 | None | None | 2025-01-14 21:01:20.570280+00:00 | 1 | None | 1 |
| 4 | missed_cleavages | int | None | False | 1 | None | None | 2025-01-14 21:01:20.570216+00:00 | 1 | None | 1 |
| 3 | enzyme_regex | str | None | False | 1 | None | None | 2025-01-14 21:01:20.570118+00:00 | 1 | None | 1 |
| 2 | amino_acid | str | None | False | 1 | None | None | 2025-01-14 21:01:20.570047+00:00 | 1 | None | 1 |
| 1 | input_dir | str | None | False | 1 | None | None | 2025-01-14 21:01:20.569827+00:00 | 1 | None | 1 |
ParamValue
| value | hash | space_id | param_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| 1 | ./fasta | None | 1 | 1 | 2025-01-14 21:01:22.677237+00:00 | 1 | None | 1 |
| 2 | C | None | 1 | 2 | 2025-01-14 21:01:22.677304+00:00 | 1 | None | 1 |
| 3 | [KR] | None | 1 | 3 | 2025-01-14 21:01:22.677351+00:00 | 1 | None | 1 |
| 4 | 0 | None | 1 | 4 | 2025-01-14 21:01:22.677404+00:00 | 1 | None | 1 |
| 5 | 4 | None | 1 | 5 | 2025-01-14 21:01:22.677455+00:00 | 1 | None | 1 |
| 6 | 75 | None | 1 | 6 | 2025-01-14 21:01:22.677503+00:00 | 1 | None | 1 |
| 7 | Executor.default | None | 1 | 7 | 2025-01-14 21:01:22.677549+00:00 | 1 | None | 1 |
Run
| uid | name | started_at | finished_at | reference | reference_type | _is_consecutive | _status_code | space_id | transform_id | report_id | _logfile_id | environment_id | initiated_by_run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | EDruK7UznZesaE6eb44l | None | 2025-01-14 21:01:21.265278+00:00 | NaT | None | None | None | 0 | 1 | 1 | NaN | None | NaN | None | 2025-01-14 21:01:21.265330+00:00 | 1 | None | 1 |
| 2 | AjDvah2V8Xw60bTrOkCv | None | 2025-01-14 21:01:22.649053+00:00 | 2025-01-14 21:01:28.871947+00:00 | 70307f22-9649-4042-b6b7-e7de177ab1bf | redun_id | None | 0 | 1 | 1 | 8.0 | None | 5.0 | None | 2025-01-14 21:01:22.649097+00:00 | 1 | None | 1 |
Storage
| uid | root | description | type | region | instance_uid | space_id | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||
| 1 | x0awepL3rlqv | /home/runner/work/redun-lamin/redun-lamin/docs... | None | local | None | iQlBPgD8uaqR | 1 | None | 2025-01-14 21:00:55.230571+00:00 | 1 | None | 1 |
Transform
| uid | key | description | type | source_code | hash | reference | reference_type | space_id | _template_id | version | is_latest | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 1 | taasWKawCiNA0000 | workflow.py | None | script | """workflow.py."""\n\n# This code is a copy fr... | Lg_7dutVm-W285sKmwyz3A | https://github.com/laminlabs/redun-lamin/blob/... | url | 1 | None | None | True | 2025-01-14 21:01:21.261136+00:00 | 1 | None | 1 |
ULabel
| uid | name | is_type | description | reference | reference_type | space_id | type_id | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | JdiyQep5 | redun | None | None | None | None | 1 | None | 2 | 2025-01-14 21:01:23.915650+00:00 | 1 | None | 1 |
******************
* module: bionty *
******************
Organism
| uid | name | ontology_id | scientific_name | synonyms | description | space_id | source_id | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||
| 1 | 1dpCL6Td | human | NCBITaxon:9606 | homo_sapiens | None | None | 1 | 1 | 2 | 2025-01-14 21:01:26.250930+00:00 | 1 | None | 1 |
Protein
| uid | name | uniprotkb_id | synonyms | description | length | gene_symbol | ensembl_gene_ids | space_id | source_id | organism_id | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 4 | 3qNrC4hwnDC9 | PO5F1_HUMAN POU domain, class 5, transcription... | Q01860 | Octamer-binding protein 3|Oct-3|Octamer-bindin... | class 5, transcription factor 1 | 360 | POU5F1 | ENST00000259915.13 [Q01860-1];ENST00000376243.... | 1 | 22 | 1 | 2 | 2025-01-14 21:01:28.852370+00:00 | 1 | None | 1 |
| 3 | 38rbzWPtKmb2 | SOX2_HUMAN Transcription factor SOX-2 | P48431 | 317 | SOX2 | ENST00000325404.3; | 1 | 22 | 1 | 2 | 2025-01-14 21:01:28.004894+00:00 | 1 | None | 1 | ||
| 2 | 6ThKerPbf6DR | KLF4_HUMAN Krueppel-like factor 4 | O43474 | Epithelial zinc finger protein EZF|Gut-enriche... | 513 | KLF4 | ENST00000374672.5 [O43474-1]; | 1 | 22 | 1 | 2 | 2025-01-14 21:01:27.149445+00:00 | 1 | None | 1 | |
| 1 | 36jnmKHdiT9m | MYC_HUMAN Myc proto-oncogene protein | P01106 | Class E basic helix-loop-helix protein 39|bHLH... | 454 | MYC | ENST00000377970.6 [P01106-1];ENST00000524013.2... | 1 | 22 | 1 | 2 | 2025-01-14 21:01:26.259675+00:00 | 1 | None | 1 |
Source
| uid | entity | organism | name | in_db | currently_used | description | url | md5 | source_website | space_id | dataframe_artifact_id | version | run_id | created_at | created_by_id | _aux | _branch_code | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 103 | 5JnV | BioSample | all | ncbi | False | True | NCBI BioSample attributes | s3://bionty-assets/df_all__ncbi__2023-09__BioS... | 918db9bd1734b97c596c67d9654a4126 | https://www.ncbi.nlm.nih.gov/biosample/docs/at... | 1 | None | 2023-09 | None | 2025-01-14 21:00:55.568608+00:00 | 1 | None | 1 |
| 102 | MJRq | bionty.Ethnicity | human | hancestro | False | True | Human Ancestry Ontology | https://github.com/EBISPOT/hancestro/raw/3.0/h... | 76dd9efda9c2abd4bc32fc57c0b755dd | https://github.com/EBISPOT/hancestro | 1 | None | 3.0 | None | 2025-01-14 21:00:55.568526+00:00 | 1 | None | 1 |
| 101 | 6vJm | bionty.DevelopmentalStage | mouse | mmusdv | False | False | Mouse Developmental Stages | http://aber-owl.net/media/ontologies/MMUSDV/9/... | 5bef72395d853c7f65450e6c2a1fc653 | https://github.com/obophenotype/developmental-... | 1 | None | 2020-03-10 | None | 2025-01-14 21:00:55.568446+00:00 | 1 | None | 1 |
| 100 | 10va | bionty.DevelopmentalStage | mouse | mmusdv | False | True | Mouse Developmental Stages | https://github.com/obophenotype/developmental-... | https://github.com/obophenotype/developmental-... | 1 | None | 2024-05-28 | None | 2025-01-14 21:00:55.568365+00:00 | 1 | None | 1 | |
| 99 | 7Zm9 | bionty.DevelopmentalStage | human | hsapdv | False | False | Human Developmental Stages | http://aber-owl.net/media/ontologies/HSAPDV/11... | 52181d59df84578ed69214a5cb614036 | https://github.com/obophenotype/developmental-... | 1 | None | 2020-03-10 | None | 2025-01-14 21:00:55.568284+00:00 | 1 | None | 1 |
| 98 | 1GbF | bionty.DevelopmentalStage | human | hsapdv | False | True | Human Developmental Stages | https://github.com/obophenotype/developmental-... | https://github.com/obophenotype/developmental-... | 1 | None | 2024-05-28 | None | 2025-01-14 21:00:55.568201+00:00 | 1 | None | 1 | |
| 97 | 1atB | Drug | all | chebi | False | False | Chemical Entities of Biological Interest | s3://bionty-assets/df_all__chebi__2024-07-27__... | https://www.ebi.ac.uk/chebi/ | 1 | None | 2024-07-27 | None | 2025-01-14 21:00:55.568107+00:00 | 1 | None | 1 |
Delete the test instance:
!rm -rf test-redun-lamin
!lamin delete --force test-redun-lamin
Show code cell output
• deleting instance testuser1/test-redun-lamin


